RESEARCH
Current Research
Biological systems follow the same physical laws as all other matter in the known universe. Yet, the interplay between innumerable coupled degrees of freedom in the condensed phase results in astounding complexity in the interwoven processes that enable life.
In the Swanson group, we use multiscale simulations to probe the mechanisms and driving forces behind biomolecular processes such as electrochemical charge transport, protein-membrane targeting, lasso peptide folding and bacterial methane oxidation. We are motivated to enable climate, medical, and societal solutions by advancing fundamental knowledge and insight.
We develop and employ a variety of multiscale methods, including ab initio, reactive and classical molecular dynamics simulations; enhanced free energy sampling; and multiscale kinetic modeling. This allows us to probe processes across a range of time and length scales, and ultimately reveal mechanistic insight. We are also actively developing multiscale kinetic modeling methods to characterize the role of kinetic selection and mechanisms of electrochemically driven transport.
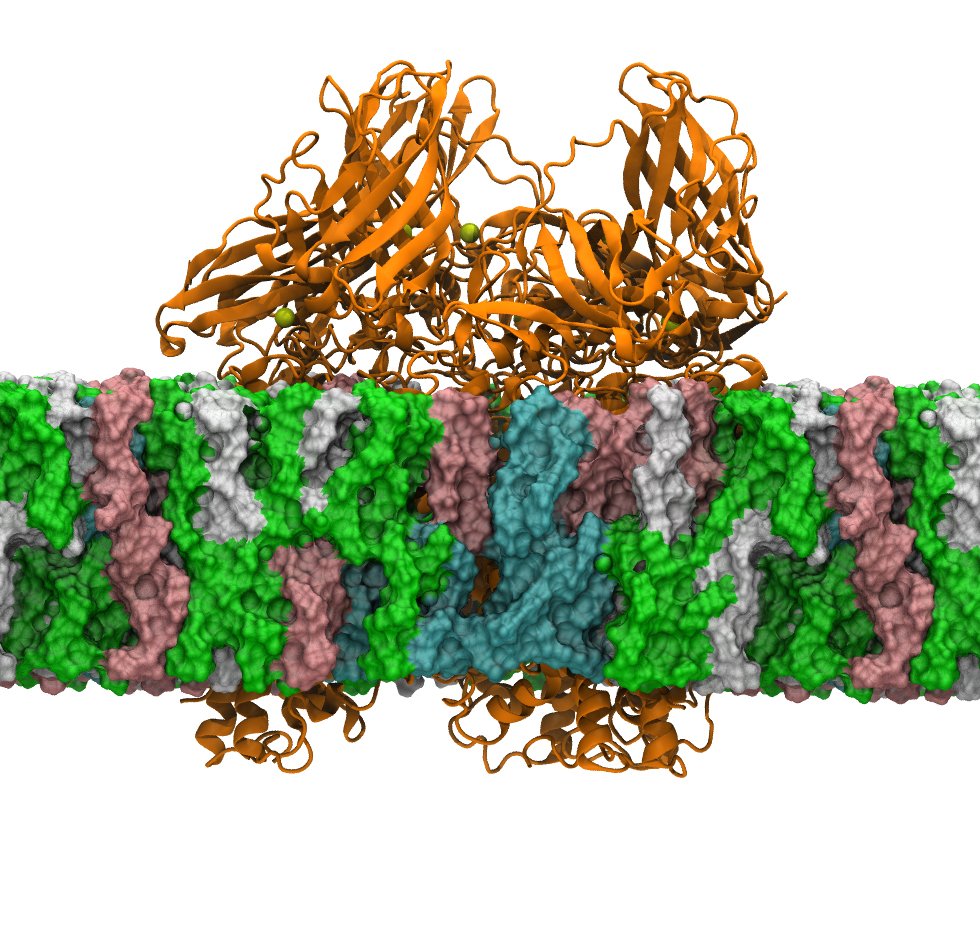
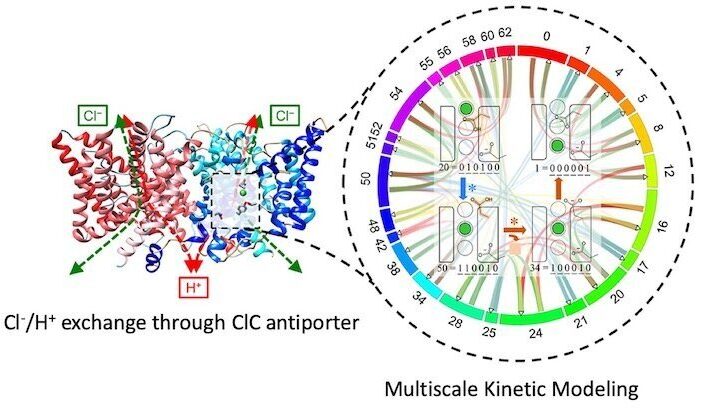
Multiscale Kinetic Modeling
Just like competing reactions in an organic synthesis, most biological processes involve multiple mechanistic pathways. Although hidden in macroscopic analyses, these competing pathways can be essential to explain and control mechanistic outcomes.
We are developing a multiscale kinetic modeling framework that combines the bottom-up quantification of rare-event transition rates with top-down refinement with experimental data to map out mechanistic reaction networks and quantify the flux through them. This provides a new level of insight into a reaction mechanism and how it changes as a function of reaction conditions.
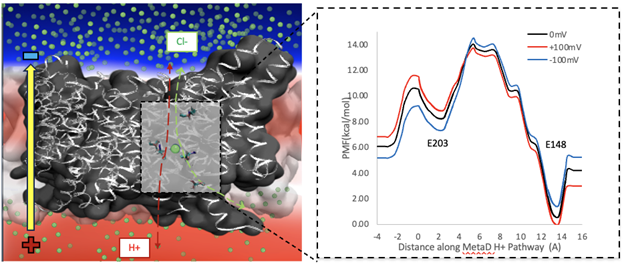
Electrochemically Driven Membrane Transport
Both passive channels and secondary active transporters control the movement of charged species across membranes in response to electrochemical potentials. Although the two driving forces—one due to a chemical gradient and the other due to a charge gradient—can be related via their Nernstian magnitudes, they are fundamentally different. We are using multiscale kinetic modeling to reveal how charge transport mechanisms can differ substantially when driven chemically and electrically. These methods explain processes like coupled ion exchange in chloride channel antiporters.
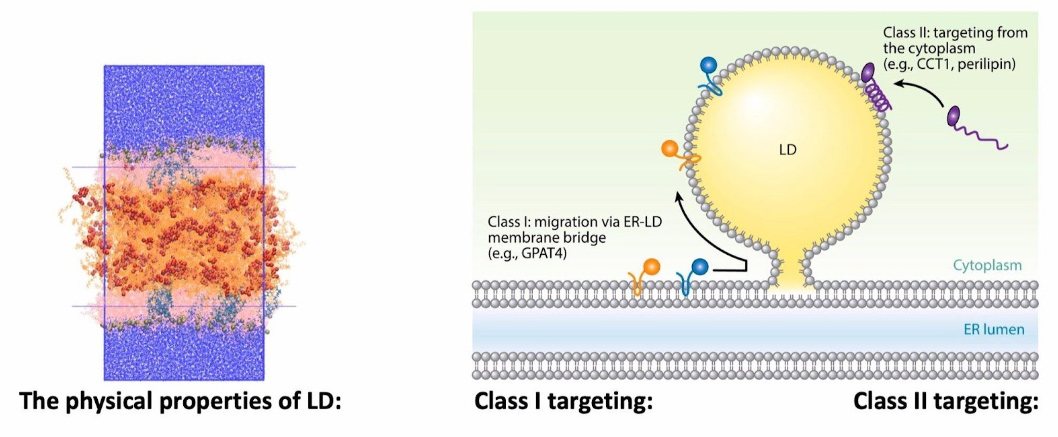
Protein-Membrane Targeting to Lipid Droplets
Lipid droplets are fascinating organelles present in all eukaryotic cells and responsible for the storage and controlled use of neutral fatty acids (fats). Since they are composed of hydrophobic molecules, they are surrounding by a phospholipid monolayer. We are using all atom simulations, protein structure prediction, enhanced sampling and other tricks to study how these monolayers are different from bilayers and how proteins specifically target lipid droplets over other organelles with the same phospholipid composition.
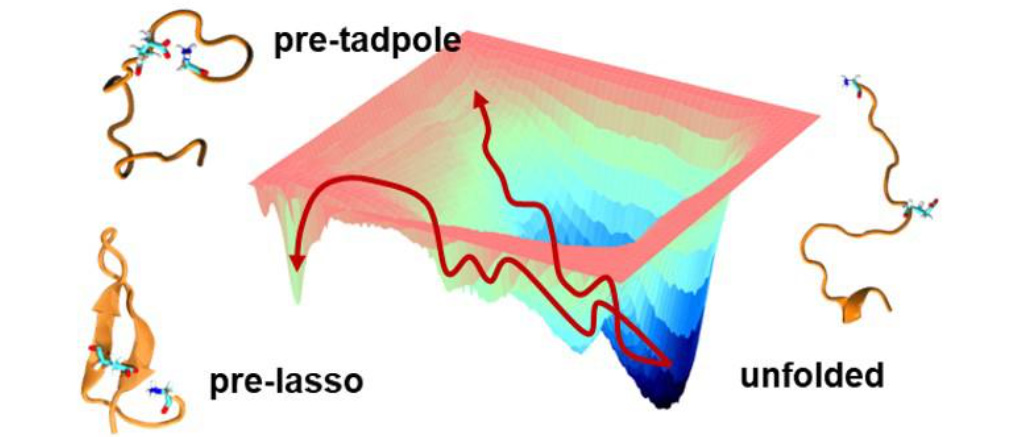
Lasso Peptide Folding
Lasso peptides are natural products with high thermal and proteolytic stabilities due to their knotted structure. Nature uses these lasso peptides as antibacterial, anticancer, and antiviral agents. We have been collaborating with the Roberts Group at the University of Utah to help them design a synthetic route to lasso peptides. If successful, non-native moieties could be added to the lasso architecture and a new class of tunable biologics with ‘mini- protein’ like properties could be accessed.
We are using enhanced sampling and protein design to characterize the lasso folding landscape and identify strategies for increasing the stability of the pre-lasso conformation, an essential prerequisite to synthesis. Our results to date demonstrate that sequence and auxiliary modifications along with reaction conditions are viable paths for stabilizing the pre-lasso motif.